双树复小波基础(Python)
双树复小波基础(Python)
·
import matplotlib.pyplot as plt
import pywtPlot Scaling and Wavelet functions for the Wavelets
w = pywt.Wavelet('db4')
print("Wavelet", w)
print("Filter bank", w.filter_bank)
print("dec_lo", w.dec_lo)
print("dec_hi", w.dec_hi)
print("rec_lo", w.rec_lo)
print("rec_hi", w.rec_hi)
# plot the decomposition and reconstruction filters
plt.figure(figsize=(12, 4))
plt.subplot(121)
plt.stem(w.dec_lo, label="Decomposition Low Pass", linefmt="-r")
plt.stem(w.dec_hi, label="Decomposition High Pass")
plt.legend()
plt.title("Decomposition Filters")
plt.grid(True)
plt.subplot(122)
plt.stem(w.rec_lo, label="Reconstruction Low Pass", linefmt="-r")
plt.stem(w.rec_hi, label="Reconstruction High Pass")
plt.legend()
plt.title("Reconstruction Filters")
plt.grid(True)
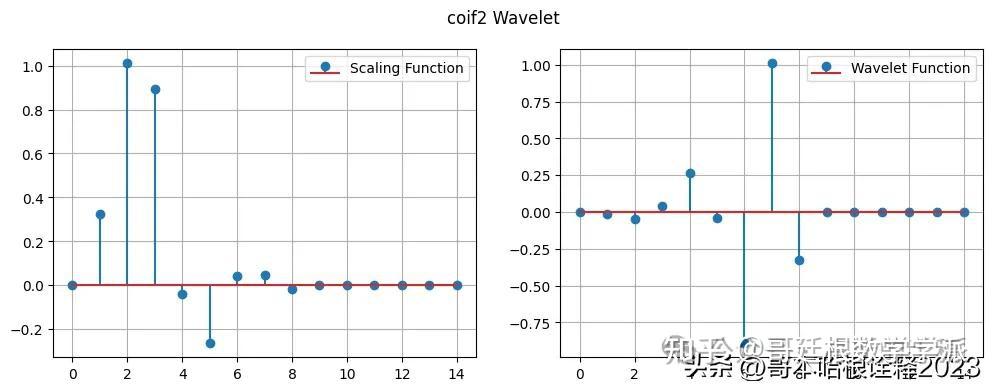
phi, psi, x = w.wavefun(level=1)
print("Scaling Coefficients", phi)
print("Detail Coefficients", psi)
print("Number of vanishing moments in detail", w.vanishing_moments_psi)
print("Number of vanishing moments in scale", w.vanishing_moments_phi)
# plot the wavelet as stem plot with linespace
plt.figure(figsize=(12, 4))
plt.subplot(121)
plt.stem(phi, label="Scaling Function")
plt.grid()
plt.legend()
plt.subplot(122)
plt.stem(psi, label="Wavelet Function")
plt.legend()
plt.suptitle("coif2 Wavelet")
plt.grid(True)
Wavelet Wavelet db4
Family name: Daubechies
Short name: db
Filters length: 8
Orthogonal: True
Biorthogonal: True
Symmetry: asymmetric
DWT: True
CWT: False
Filter bank ([-0.010597401785069032, 0.0328830116668852, 0.030841381835560764, -0.18703481171909309, -0.027983769416859854, 0.6308807679298589, 0.7148465705529157, 0.2303778133088965], [-0.2303778133088965, 0.7148465705529157, -0.6308807679298589, -0.027983769416859854, 0.18703481171909309, 0.030841381835560764, -0.0328830116668852, -0.010597401785069032], [0.2303778133088965, 0.7148465705529157, 0.6308807679298589, -0.027983769416859854, -0.18703481171909309, 0.030841381835560764, 0.0328830116668852, -0.010597401785069032], [-0.010597401785069032, -0.0328830116668852, 0.030841381835560764, 0.18703481171909309, -0.027983769416859854, -0.6308807679298589, 0.7148465705529157, -0.2303778133088965])
dec_lo [-0.010597401785069032, 0.0328830116668852, 0.030841381835560764, -0.18703481171909309, -0.027983769416859854, 0.6308807679298589, 0.7148465705529157, 0.2303778133088965]
dec_hi [-0.2303778133088965, 0.7148465705529157, -0.6308807679298589, -0.027983769416859854, 0.18703481171909309, 0.030841381835560764, -0.0328830116668852, -0.010597401785069032]
rec_lo [0.2303778133088965, 0.7148465705529157, 0.6308807679298589, -0.027983769416859854, -0.18703481171909309, 0.030841381835560764, 0.0328830116668852, -0.010597401785069032]
rec_hi [-0.010597401785069032, -0.0328830116668852, 0.030841381835560764, 0.18703481171909309, -0.027983769416859854, -0.6308807679298589, 0.7148465705529157, -0.2303778133088965]
Scaling Coefficients [ 0. 0.32580343 1.01094572 0.89220014 -0.03957503 -0.26450717
0.0436163 0.0465036 -0.01498699 0. 0. 0.
0. 0. 0. ]
Detail Coefficients [ 0. -0.01498699 -0.0465036 0.0436163 0.26450717 -0.03957503
-0.89220014 1.01094572 -0.32580343 0. 0. 0.
0. 0. 0. ]
Number of vanishing moments in detail 4
Number of vanishing moments in scale 0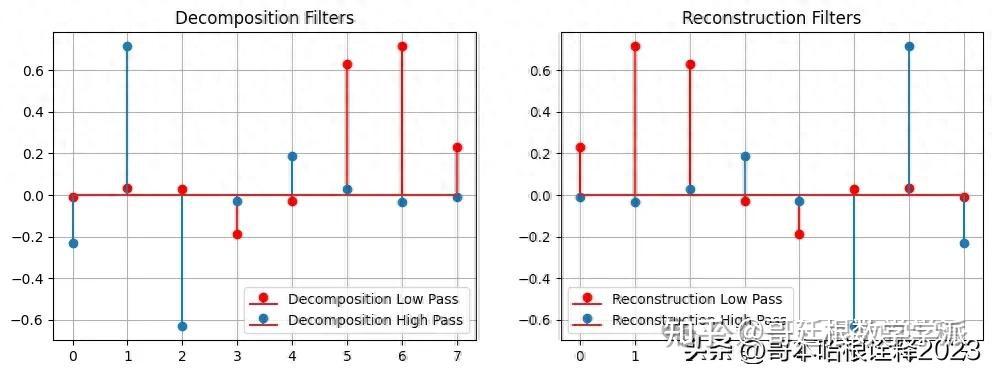
Visualizing 2D Wavelets
import numpy as np
import pywt
import matplotlib.pyplot as plt
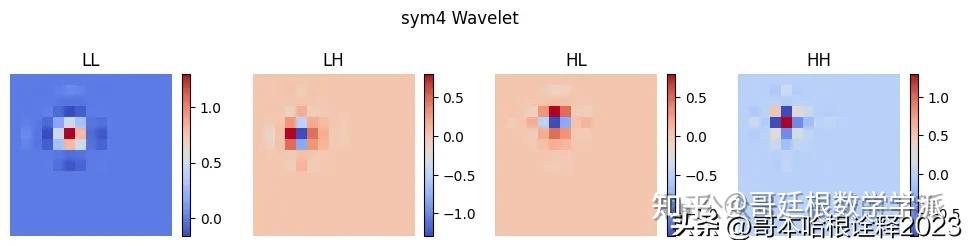
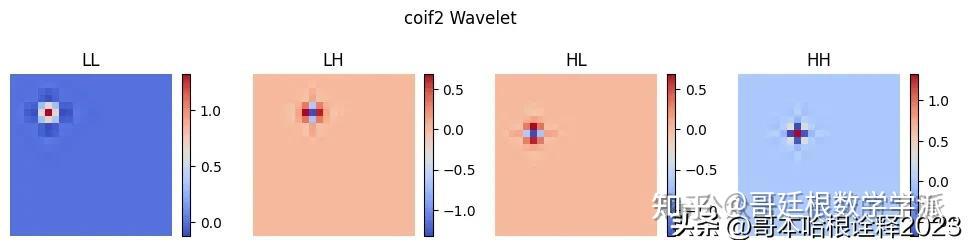
for type in ['haar', 'db4', 'sym4', 'coif2']:
wavelet = pywt.Wavelet(type)
phi, psi, x = wavelet.wavefun(level=1)
# Create a 2D grid
xx, yy = np.meshgrid(x, x)
labels = ["LL", "LH", "HL", "HH"]
plt.figure(figsize=(12, 3))
i = 0
for s in [phi, psi]:
for w in [phi, psi]:
# Calculate 2D wavelet
wavelet = np.outer(s, w)
plt.subplot(1, 4, i + 1)
plt.imshow(wavelet, cmap='coolwarm', extent=[x.min(), x.max(), x.min(), x.max()])
plt.title(labels[i])
plt.xlabel('X')
plt.colorbar(shrink=0.7)
plt.ylabel('Y')
plt.axis('off')
# # Plot the 3D surface
# ax = plt.subplot(2, 4, i+5, projection='3d')
# surf = ax.plot_surface(xx, yy, wavelet, cmap='coolwarm')
# # ax.set_title(labels[i])
# ax.set_xlabel('X')
# ax.set_ylabel('Y')
# ax.set_zlabel('Amplitude')
# plt.colorbar(surf, shrink=0.5, aspect=5, label='Amplitude')
i += 1
plt.suptitle(f"{type} Wavelet")
plt.show()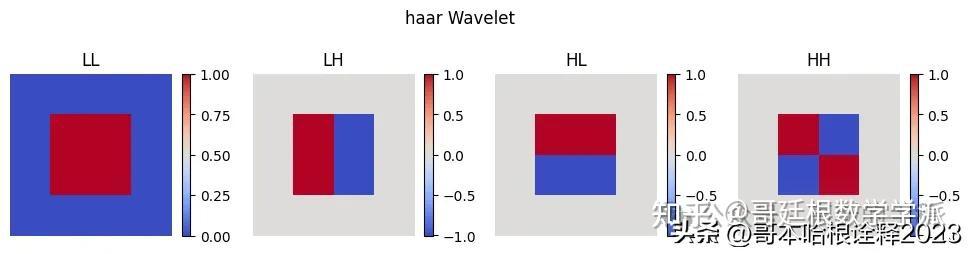
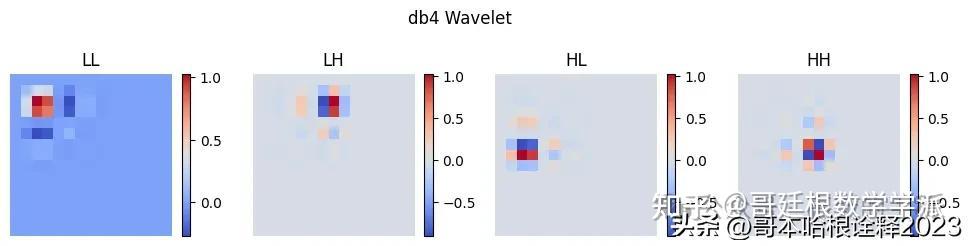
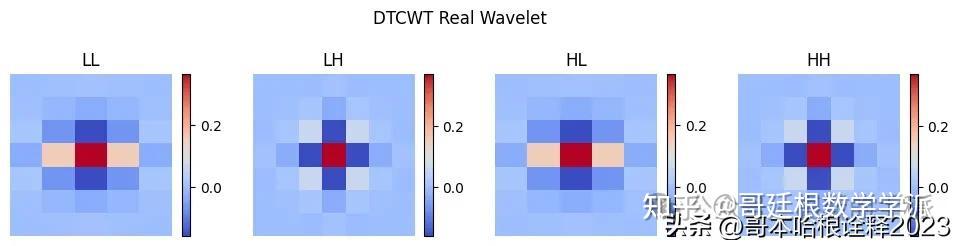
import dtcwt.coeffs
coeefs = dtcwt.coeffs.biort('near_sym_a') # h0, g0, h1, g1
phi0, phi1, psi0, psi1 = [c.reshape(-1,) for c in coeefs]
labels = ["LL", "LH", "HL", "HH"]
tree_a = []
plt.figure(figsize=(12, 3))
i = 0
for s in [psi0, psi0]:
for w in [phi0, psi0]:
# Calculate 2D wavelet
wavelet = np.outer(s, w)
print(wavelet.shape)
tree_a.append(wavelet)
plt.subplot(1, 4, i + 1)
plt.imshow(wavelet, cmap='coolwarm', extent=[x.min(), x.max(), x.min(), x.max()])
plt.title(labels[i])
plt.xlabel('X')
plt.colorbar(shrink=0.7)
plt.ylabel('Y')
plt.axis('off')
i += 1
plt.suptitle(f"DTCWT Real Wavelet")
plt.show()
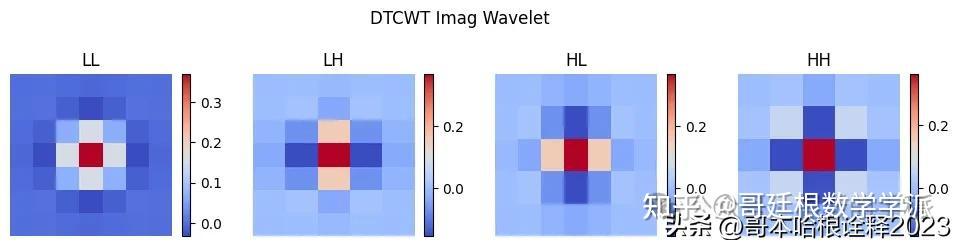
tree_b = []
plt.figure(figsize=(12, 3))
i = 0
for s in [phi1, psi1]:
for w in [phi1, psi1]:
# Calculate 2D wavelet
wavelet = np.outer(s, w)
print(wavelet.shape)
tree_b.append(wavelet)
plt.subplot(1, 4, i + 1)
plt.imshow(wavelet, cmap='coolwarm', extent=[x.min(), x.max(), x.min(), x.max()])
plt.title(labels[i])
plt.xlabel('X')
plt.colorbar(shrink=0.7)
plt.ylabel('Y')
plt.axis('off')
i += 1
plt.suptitle(f"DTCWT Imag Wavelet")
plt.show()
# Wavelts
plt.figure(figsize=(12, 6))
i = 0
for s in [phi1, psi1]:
for w in [phi1, psi1]:
# Calculate 2D wavelet
wavelet1 = tree_a[i] + tree_b[3 - i]
wavelet2 = tree_a[i] - tree_b[3 - i]
plt.subplot(2, 4, i + 1)
plt.imshow(wavelet1, cmap='coolwarm', extent=[x.min(), x.max(), x.min(), x.max()])
plt.title(labels[i])
plt.xlabel('X')
plt.colorbar(shrink=0.7)
plt.ylabel('Y')
plt.axis('off')
plt.subplot(2, 4, i + 5)
plt.imshow(wavelet2, cmap='coolwarm', extent=[x.min(), x.max(), x.min(), x.max()])
plt.title(labels[i])
plt.xlabel('X')
plt.colorbar(shrink=0.7)
plt.ylabel('Y')
plt.axis('off')
i += 1
plt.suptitle(f"DTCWT Wavelets")
plt.show()
# Load the mandrill image
mandrill = plt.imread("../star.png")
mandrill = mandrill.mean(axis=2)
# Show mandrill
plt.figure(1)
plt.axis('off')
plt.title("Sample Image")
plt.imshow(mandrill, cmap='gray', clim=(0,1))
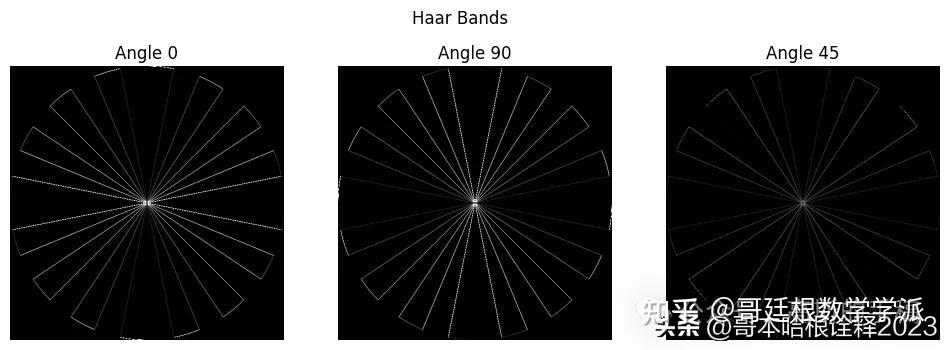
LL, wavelets_haar = pywt.dwt2(mandrill, 'haar')
print(len(wavelets_haar))
# Show the absolute images for each direction in level 2.
# Note that the 2nd level has index 1 since the 1st has index 0.
labels = [0, 90, 45]
plt.figure(2, figsize=(12, 4))
plt.suptitle("Haar Bands")
for slice_idx in range(len(wavelets_haar)):
plt.subplot(1, 3, slice_idx + 1)
plt.axis('off')
plt.title(f"Angle {labels[slice_idx]}")
plt.imshow(10 * (np.abs(wavelets_haar[slice_idx])), cmap="gray", clim=(0, 1))
DTCWT Demonstration
# Load the mandrill image
mandrill = plt.imread("../star.png")
mandrill = mandrill.mean(axis=2)
# Show mandrill
plt.figure(1)
plt.axis('off')
plt.title("Sample Image")
plt.imshow(mandrill, cmap='gray', clim=(0,1))
import dtcwt
import numpy as np
transform = dtcwt.Transform2d()
# Compute two levels of dtcwt with the defaul wavelet family
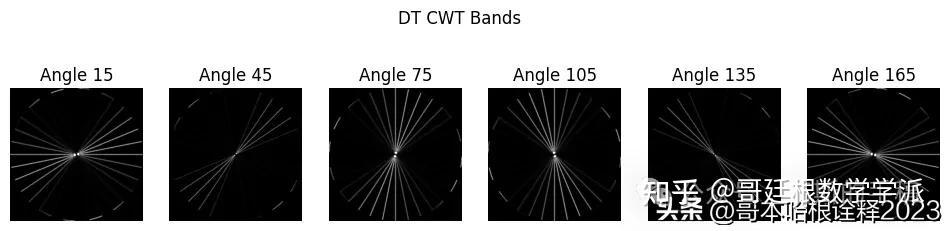
mandrill_t = transform.forward(mandrill, nlevels=2)
# Show the absolute images for each direction in level 2.
# Note that the 2nd level has index 1 since the 1st has index 0.
labels = [15, 45, 75, 105, 135, 165]
plt.figure(2, figsize=(12, 3))
plt.suptitle("DT CWT Bands")
for slice_idx in range(mandrill_t.highpasses[1].shape[2]):
plt.subplot(1, 6, slice_idx + 1)
plt.axis('off')
plt.title(f"Angle {labels[slice_idx]}")
plt.imshow(5 * (np.abs(mandrill_t.highpasses[1][:,:,slice_idx])), cmap="gray", clim=(0, 1))
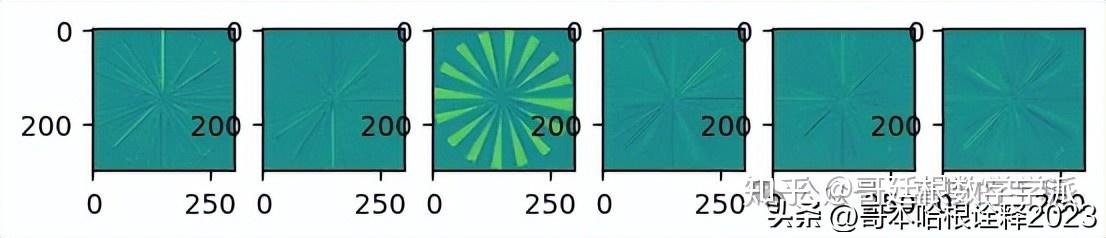
# Show the phase images for each direction in level 2.
plt.figure(3)
for slice_idx in range(mandrill_t.highpasses[1].shape[2]):
plt.subplot(1, 6, slice_idx + 1)
plt.imshow(np.angle(mandrill_t.highpasses[1][:,:,slice_idx]), cmap="viridis", clim=(-np.pi, np.pi))
import numpy as np
from scipy.signal import hilbert
# Input signal
signal = np.array([-0.05, 0.25, 0.6, 0.25, -0.05])
# Compute the Hilbert Transform
analytic_signal = hilbert(signal)
hilbert_transform = np.imag(analytic_signal)
# Display the results
print("Original Signal: ", signal)
print("Hilbert Transform: ", hilbert_transform)
Original Signal: [-0.05 0.25 0.6 0.25 -0.05]
Hilbert Transform: [-1.33803035e-01 -3.56506308e-01 2.22044605e-17 3.56506308e-01
1.33803035e-01]
plt.figure(figsize=(12, 8))
plt.subplot(2, 2, 1)
plt.stem(h0)
plt.title("Analysis H0")
plt.subplot(2, 2, 2)
plt.stem(h1)
plt.title("Analysis H1")
plt.subplot(2, 2, 3)
plt.stem(g0)
plt.title("Recon G0")
plt.subplot(2, 2, 4)
plt.stem(g1)
plt.title("Recon G1")
plt.show()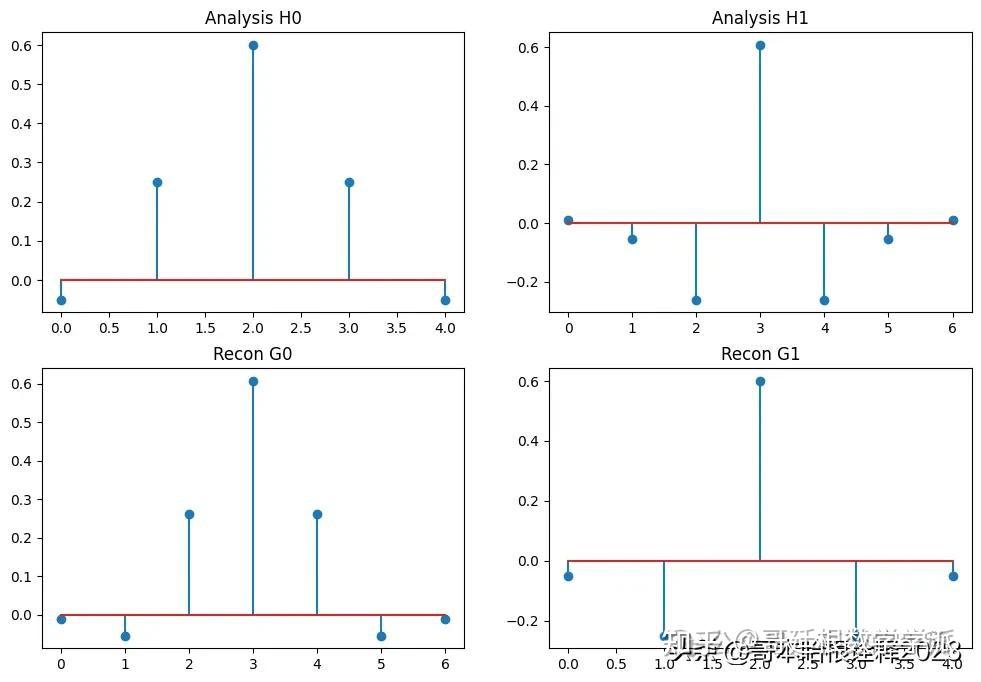
plt.figure(figsize=(12, 8))
plt.subplot(2, 2, 1)
a = np.asarray([0] + list(h0.reshape(-1, )) + [0])
b = h1.reshape(-1)
plt.stem(a + b)
plt.title("Adding 5 tap LP and 7 tap HP")
plt.subplot(2, 2, 2)
plt.plot(a + b)
plt.title("Adding 5 tap LP and 7 tap HP")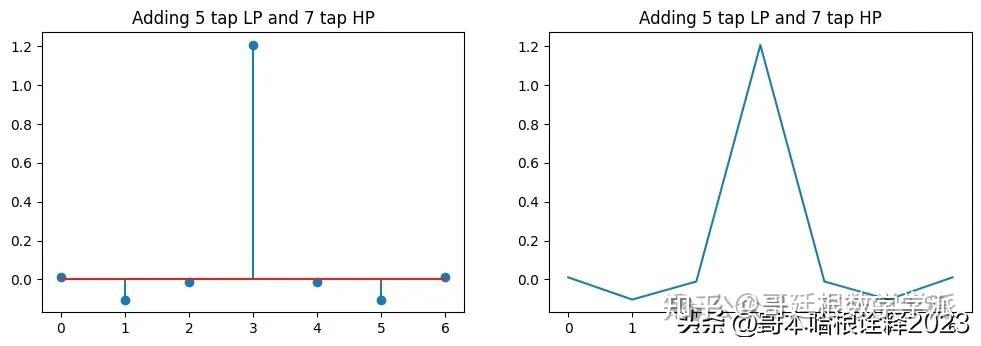
import numpy as np
import matplotlib.pyplot as plt
import dtcwt
# Function to plot the wavelets
def plot_dtcwt_wavelets(levels=3):
# Create a 1D signal
signal_length = 64
signal = np.zeros(signal_length)
signal[signal_length // 2] = 1 # Impulse signal
# Perform Dual-Tree Complex Wavelet Transform
transform = dtcwt.Transform1d()
wt = transform.forward(signal, nlevels=levels)
# Plot the real and imaginary parts of wavelets at each level
fig, axes = plt.subplots(2, levels, figsize=(15, 6))
for level in range(levels):
# Extract wavelet coefficients
wavelet_coeffs = wt.highpasses[level]
x = np.arange(len(wavelet_coeffs))
# Plot real part
axes[0, level].plot(x, wavelet_coeffs.real, label='Real', color='blue')
axes[0, level].set_title(f'Level {level + 1} - Real Part')
axes[0, level].grid(True)
# Plot imaginary part
axes[1, level].plot(x, wavelet_coeffs.imag, label='Imaginary', color='orange')
axes[1, level].set_title(f'Level {level + 1} - Imaginary Part')
axes[1, level].grid(True)
# Add labels
axes[0, 0].set_ylabel('Amplitude')
axes[1, 0].set_ylabel('Amplitude')
axes[1, 1].set_xlabel('Index')
plt.tight_layout()
plt.show()
# Call the function to plot wavelets
plot_dtcwt_wavelets(levels=4)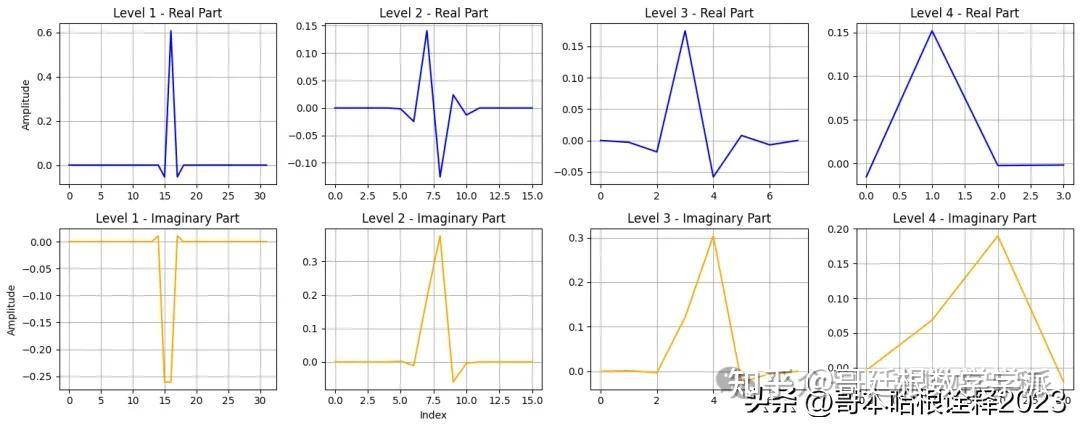
学术咨询:

担任《Mechanical System and Signal Processing》《中国电机工程学报》等期刊审稿专家,擅长领域:信号滤波/降噪,机器学习/深度学习,时间序列预分析/预测,设备故障诊断/缺陷检测/异常检测。
分割线分割线分割线
基于小波分析的Linear电磁谱降噪(Python)



Python环境下基于最小最大凹面全变分一维信号降噪方法

MATLAB环境下简单的PPG信号(光电容积脉搏波信号)分析方法(滤波降噪分解等)

基于双树复小波和邻域多尺度的非平稳信号降噪方法(MATLAB)


更多推荐
 已为社区贡献11条内容
已为社区贡献11条内容







所有评论(0)